Pivoting with Patience
Neuman Lab Looks to Examine Coronavirus Inhibitors
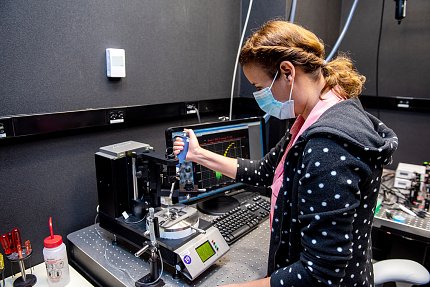
Photo: NIH Office of Human Resources
This article begins a special series on pandemic pivots—intramural scientists and labs that changed course to tackle Covid-related research.
Since 2006, Dr. Keir Neuman, senior investigator in NHLBI’s Laboratory of Single Molecule Biophysics, has studied enzymes—specifically ones that control or manipulate the structure of DNA. His group has had enormous success with two in particular—topoisomerases, which untwist and untangle DNA, and helicases, which unwind the double helix.
Pre-Covid and extreme telework, you could find several members of the team in the ultra-quiet darkness of a custom-designed lab, specially calibrated for light, sound and motion. Scientists who came to biology by way of physics, they might look like they are adding super-tiny Lego pieces to a desk-size 3-D puzzle. In a sense, they are.
“We’re trying to measure the motion of something that’s about 100,000 times less than the width of a human hair,” Neuman said, describing the individual molecules they’re stalking. “That’s what we’re trying to measure directly. So, this is a very precise, very stable kind of work. And it’s all done in real time. This is what’s really beautiful. We watch a single piece of DNA.”
The scientists use instruments they build themselves by combining micro-tweezers, video cameras, desktop computers and the software that operates it all. They attach a small magnet to the DNA molecule and then watch as the protein changes the structure of the DNA.
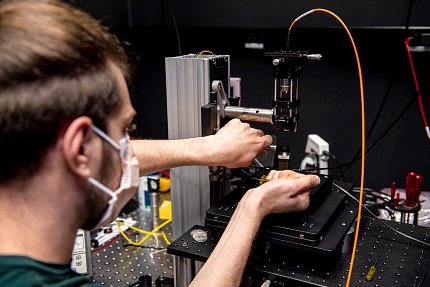
Photo: NIH Office of Human Resources
“We see even small, minor moves—up and down,” Neuman said. “We measure and can respond to that in real time. We can see by eye a single enzyme working. And there’s something incredibly compelling about that.”
When Covid struck in early 2020, the team recently had witnessed significant milestones. They’d seen how solo topoisomerases relaxing overwound DNA might behave when faced with a virtual stop sign—a chemotherapeutic drug. Neuman pitched an idea to his team: Could they adapt their observations to potential Covid drugs that work by inhibiting viral replication?
“If we had a really good sense of the molecular basis of inhibition—why does this drug work and the next one not work?—then we could get a handle on what’s effective, and that may help in producing new drugs,” he mused.
The lab’s staff scientist and “technical wizard,” Dr. Yeonee Seol, immediately seized on the idea.
“As a scientist, particularly as a basic scientist, it’s not often you feel like you’re contributing in a direct way,” Neuman said. “I think she was really upset by what was going on and thought this would be our way to fight back.”
About the same time, NIH announced an Intramural Research Program funding competition: Who could pivot to a pandemic-related project? Neuman and Seol wrote up a proposal that became 1 of about 40 to get the green light.
“It’s a new view of looking at biology,” Neuman said. “This technique we’ve developed is highly applicable to this situation and has the potential to give us unique insights into how this process of viral RNA replication takes place.”
Consider watching 100,000 people gathered in a room, Neuman suggested. Now try to document how each one is behaving.
“You get an average,” he said, “but you don’t know the details—what [individual] people are doing or what they’re thinking. With most work on Covid, replication is done in a test tube”—hundreds of thousands of folks to watch at once. “Think of the single-molecule approach. What we realized from more than a decade of this work is that you learn an enormous amount by understanding what an individual molecule is doing.”

Photo: courtesy keir neuman
Topoisomerases turn out to be an important target for chemotherapeutic anti-cancer drugs. Neuman’s group discovered that by observing exactly how individual enzymes are inhibited, he and colleagues can get a deep appreciation for therapy development.
“We thought that by studying the replication process of Covid-19—the so-called RNA-dependent RNA polymerase, how it’s inhibited and the molecular details of how this inhibition takes place, which could be lost or missed in ensemble or traditional biochemical assays—that we may be able to shed light on this process, and help refine [inhibitor drugs such as Remdesivir]…What we do understand [about enzyme-DNA interaction] is exquisitely complex. And what we’ve learned from this technique is that it’s a way of sort of unpacking that complexity.”
With the go-ahead and funding from IRP, the lab was able to recruit Dr. Hajnalka Harami-Papp, a visiting fellow from Hungary with expertise in viral biochemistry who will focus on the replication process.
Currently several lab members are preparing substrates, making proteins and otherwise gearing up to tackle this new target in what Neuman describes as less of a full pivot and more of an expansion of their original work. Next steps involve a cross-institute collaboration.
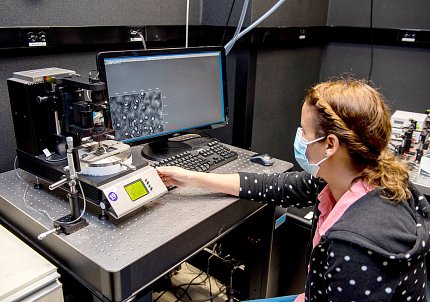
Photo: NIH Office of Human Resources
“My colleague Dr. Wei Wang is a structural biologist in NIDDK who also pivoted to some extent to study the structure, not just of the polymerase, but also as much of that complex as she possibly can,” Neuman explained. “And so, our hope is that if she’s successful in purifying the whole replication complex—with every single protein involved—then we can leverage her work and study that also at the single-molecule level.”
The hardest part of even a partial pivot for groups like Neuman’s may be adjusting expectations while confronting a crisis.
“The one trouble with a single-molecule approach is that it’s slow and we’ve all had to learn to be very, very patient,” he concluded. “The insights are spectacular, but we measure the activity of a single enzyme. We all know that anecdotes don’t carry a lot of water, right? You can see every detail in this process, but you can’t write a paper about one enzyme. We have to collect a hundred or a few hundred individual measurements—and that’s per condition. The data collection can be exceedingly slow.
“Some people in the lab get frustrated at times, because we collaborate with people who do more traditional ensemble assays,” he continued. “In an afternoon, they can test things that take us weeks to test. And yet, we have this level of detail that is very difficult to achieve with other approaches. So, we get used to things taking a while. And, from what we’ve all come to understand over the past few years is, [Covid-19] won’t be the last [infection] we’ll face. There’ll always be another one.”





